- Understand which panel makes sense for your patient.
- How is ctDNA detected from plasma?
- Understand what tumor fraction means.
- Learn how to interpret copy number data.
- Get a grasp on variant interpretation.
- Understand the latest bioinformatics behind future plasma DNA applications.
Hi there,
we're the authors of the publication below that you probably just read. As stated, we believe that especially young oncologists' careers will benefit greatly from learning about the fundamentals of genomics and liquid biopsy profiling early on. So without much further ado, let's get right to it.

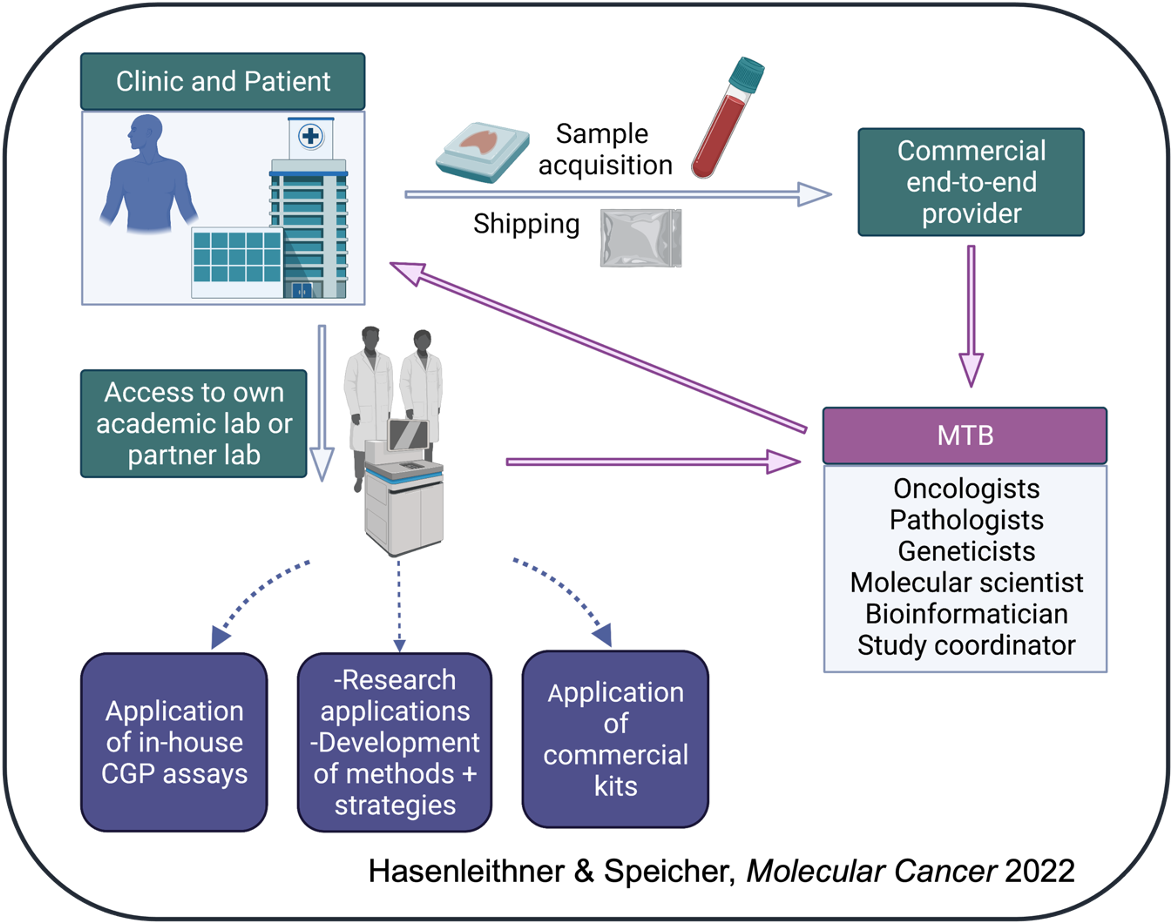
Whether ordering ctDNA testing from your local lab or an industry provider, it is important to know the steps taken before trying to implement the genomic findings into clinical care.
Vessel offers career programs and projects through wet and dry lab processes, as well as an interpretation tutorial from real-world cases studies using ctDNA profiling (Hasenleithner & Speicher, Molecular Cancer 2022).
Real-world case studies
We're going to work on two use cases for ctDNA analysis throughout the patient journey:
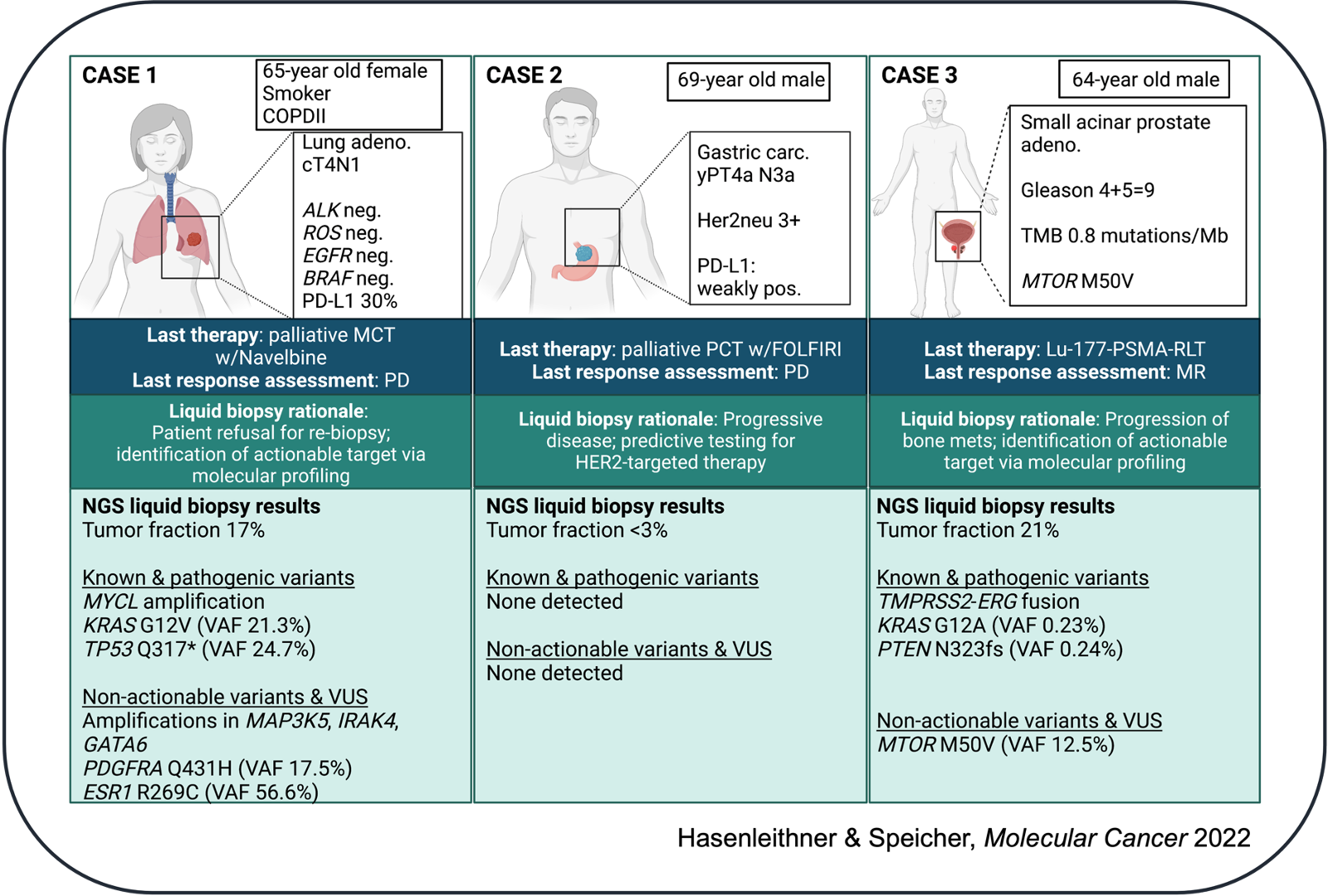
(1) Identification of actionable targets from ctDNA in patients with advanced cancer
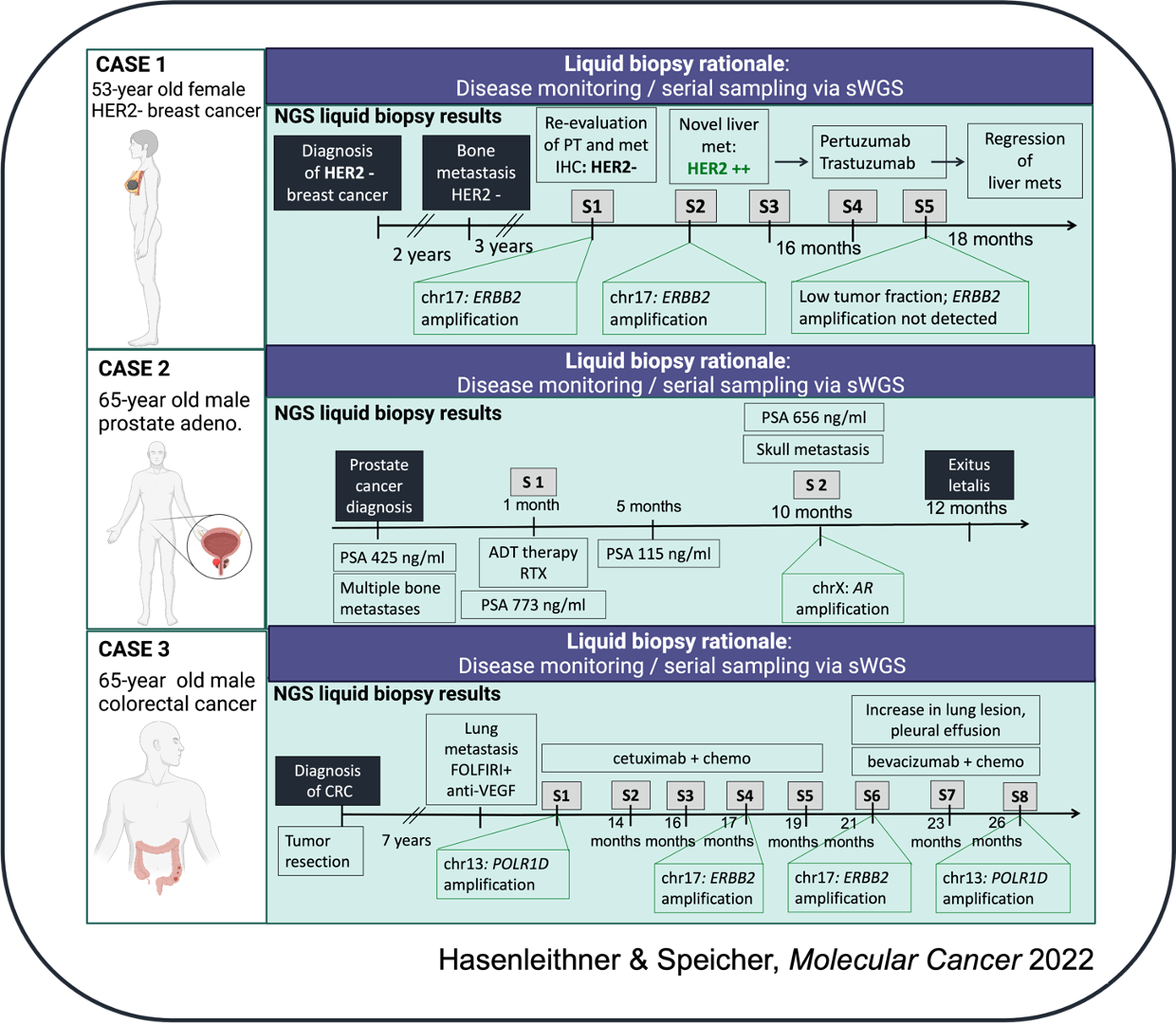
and (2) Disease monitoring
Before we dive into the real-world cases, let's take a "meta" look at the procedures involved in the interpretation of cfDNA-based data.
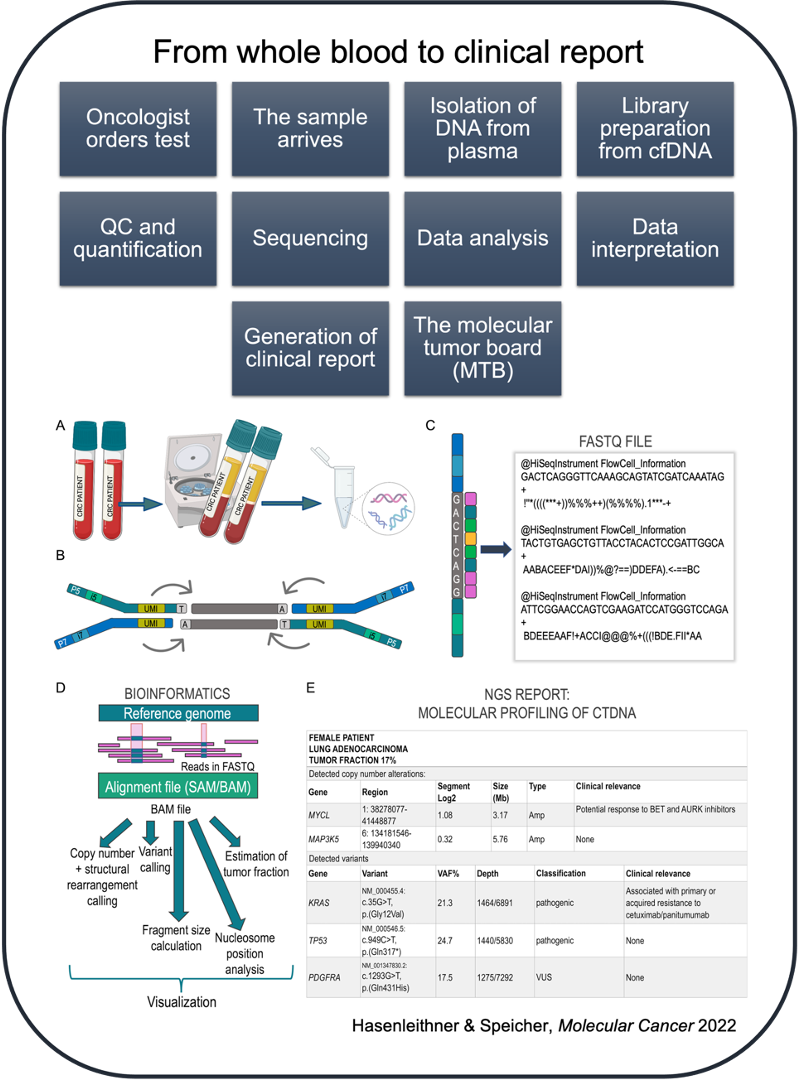
From whole blood to clinical report (a high-level overview)
There are a number of pre-analytical sampling handling factors that influence downstream analysis of cell-free DNA (cfDNA). It helps to be aware of these factors when working with cfDNA testing, as they may significantly affect the integrity, purity and yield of a sample.
Previous works have detailed the influence of pre-analytical factors, such as the choice of blood collection tubes, which may have varying stabilization reagents, sample storage conditions and duration, centrifugation steps for plasma separation, cfDNA purification, quantification and characterization, as well as library preparation protocols.

In this regard, several collaborative efforts are working to evaluate these findings, devise the criteria for standardizing liquid biopsy workflows, put these into context for providers and end users of cfDNA assays, and to bridge the gap between academia and industry to enable the translation of research findings into clinical implementation:
| Initiative | Source |
|---|---|
| CANCER-ID | https://www.cancer-id.eu/ (project completed) |
| The European Liquid Biopsy Society (ELBS) | https://www.uke.de/english/departments-institutes/institutes/tumor-biology/european-liquid-biopsy-society-elbs/index.html |
| BLOODPAC | https://www.bloodpac.org/ |
Once cfDNA has been successfully isolated and quantified, a library preparation and subsequent sequencing strategy must be selected. The strategy will vary depending on the type of ctDNA signal you are trying to measure and, of course, so too will the price of sequencing.
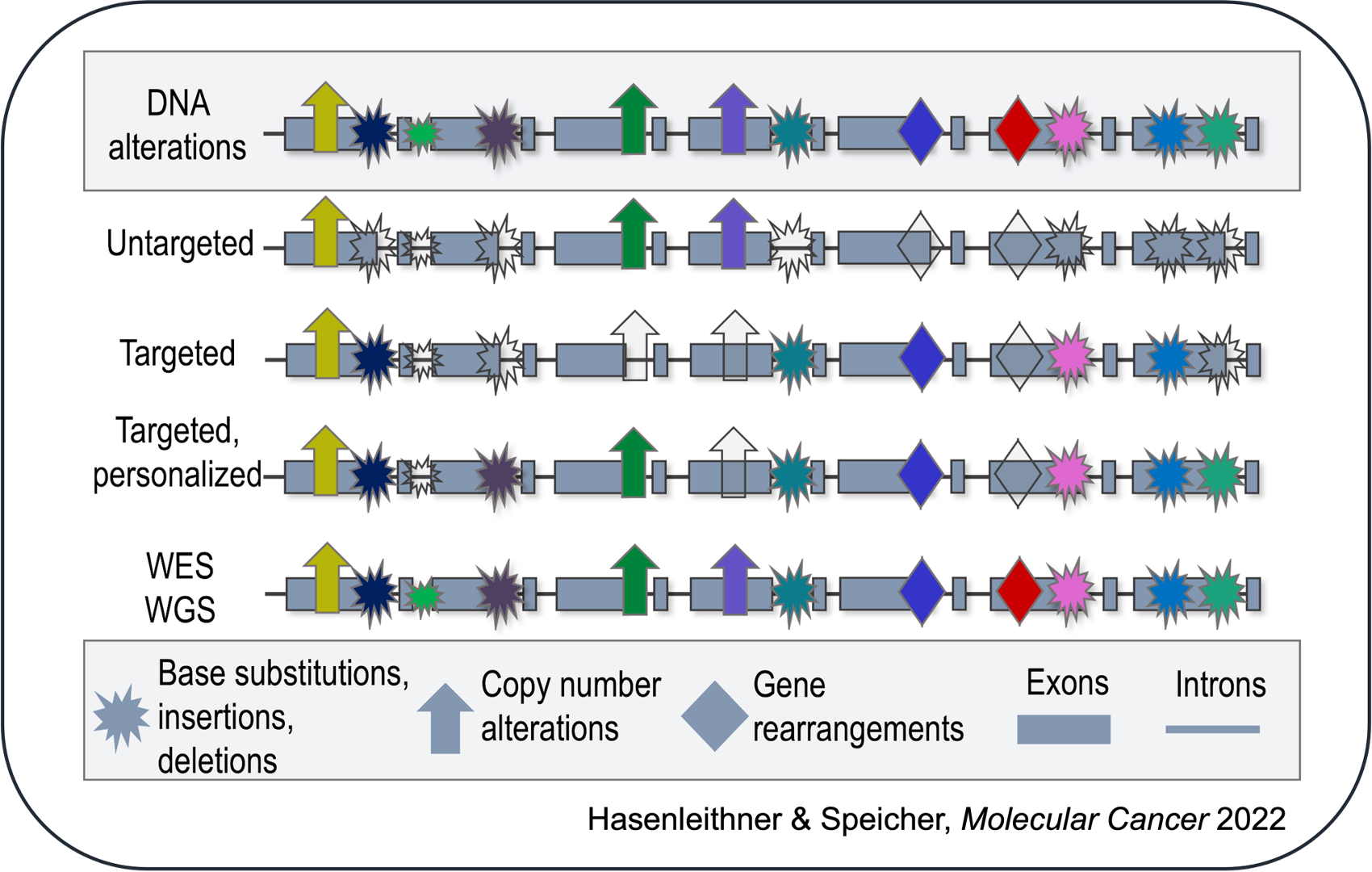
For example, are you only interested in detecting somatic copy number alterations (SCNA)? If that is the case, an untargeted, shallow whole-genome sequencing (sWGS) approach would suffice. Do you have a priori knowledge of the patient's tumor and would like to selectively track these mutations through liquid biopsy monitoring? This would indicate the use of a targeted, personalized approach. Are you looking to identify regions of open chromatin to infer tissue-of-origin of cfDNA fragments? For this purpose, a whole-genome sequencing (WGS) approach at higher coverage would be the most suitable. However, it is important to be aware that not every alteration in a patient's sample will be detected by every approach and assay sensitivity is variable.
After sequencing has been performed, imaging data from the sequencer are converted into base calls, which are then converted into sample-specific raw data. The sequencing reads and quality scores are stored in the universal FASTQ format.
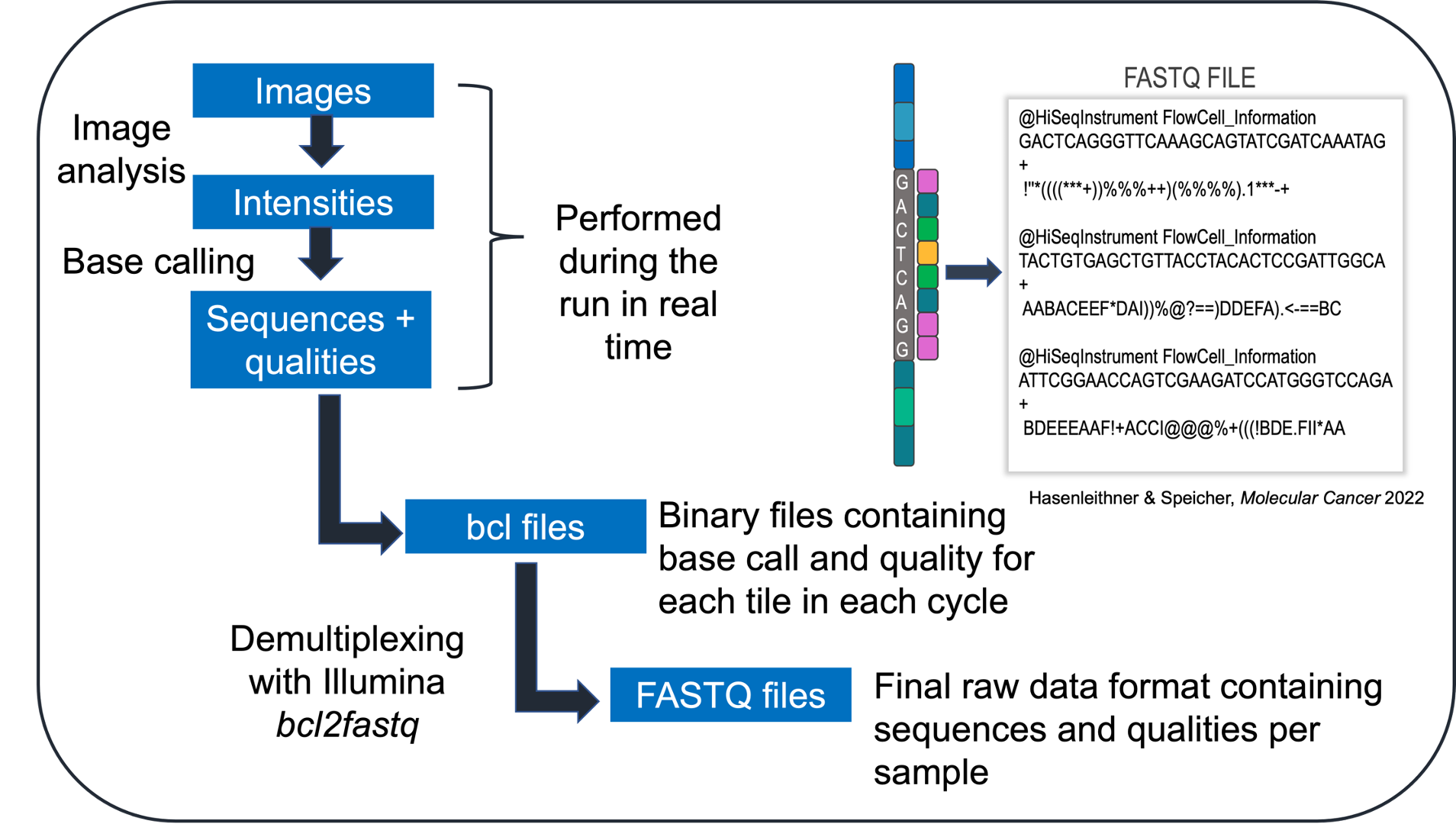
What does a FASTQ data file look like when you open it?

The FASTQ format has 4 lines of data per sequence:

The reads stored in the FASTQ file do not contain any positional information. How can we predict the locus from which the read originates? This is done through a step called read alignment, or mapping. Essentially, the reads are mapped to a reference genome using an alignment software.
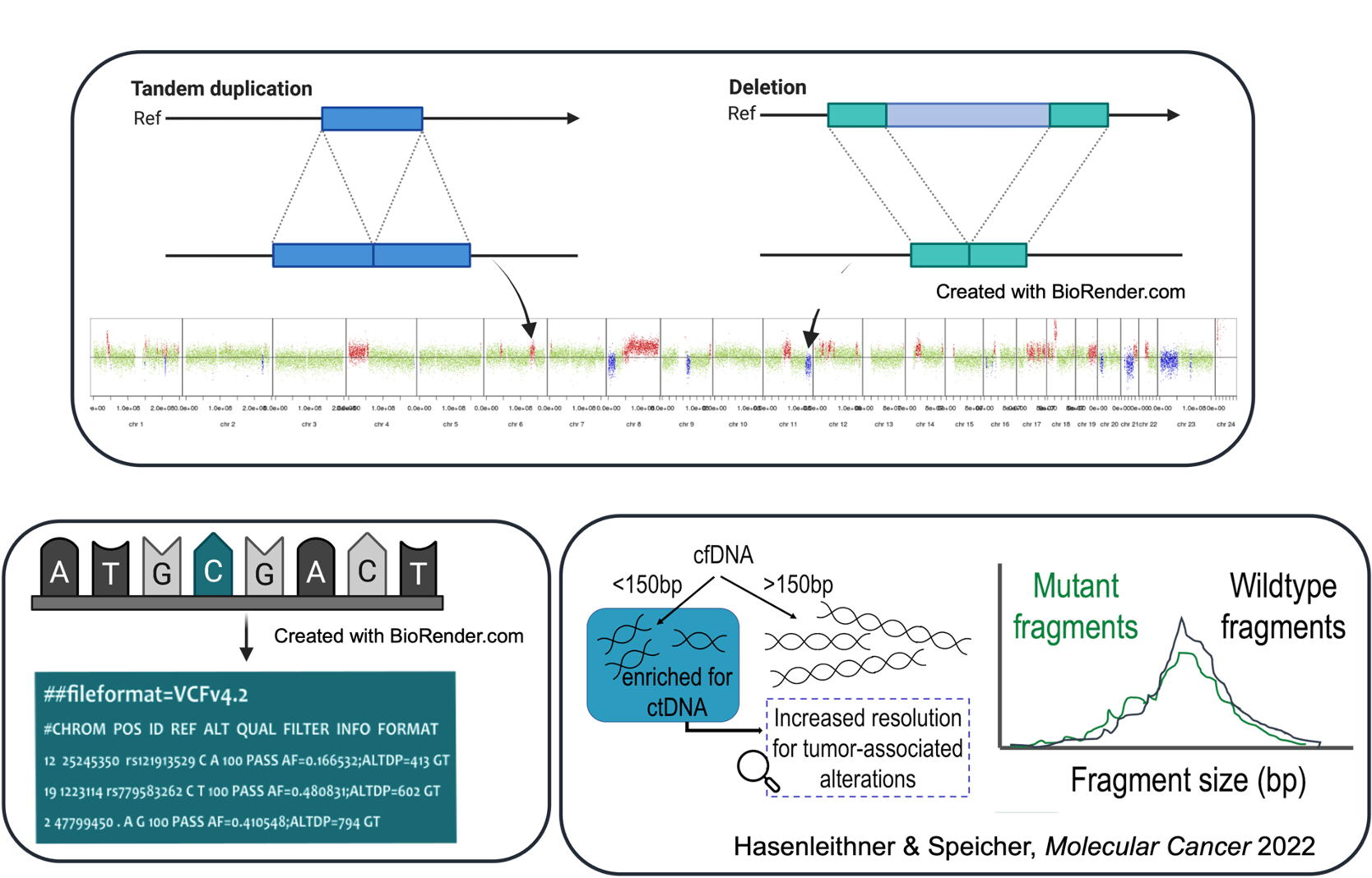
Genomic variance is derived through the comparison with the reference genome. Harvesting the ctDNA signal from the alignment data varies dramatically, depending on the sequencing strategy and clinical question, and is conducted with various computational pipelines. For example, the process of mutation detection (SNVs + InDels) is called variant calling. This differs from e.g. copy number or structural variant calling, which requires a different computational strategy. Similarly, fragmentomics-based approaches require specially tailored algorithms depending on the feature being harvested. This entire workflow of NGS data analysis, i.e. bioinformatics, is one of the most decisive aspects of ctDNA testing, yet it represents the step most lacking in transparency for clinicians. The "black box" of bioinformatics serves as the backbone for much uncertainty when it comes to understanding and trusting data generated by next-generation sequencing.
Interpretation of data from liquid biopsy is potentially the most complex step in the ctDNA testing workflow, especially as it requires expertise about bioinformatics-related limitations as well as cancer knowledgebases.
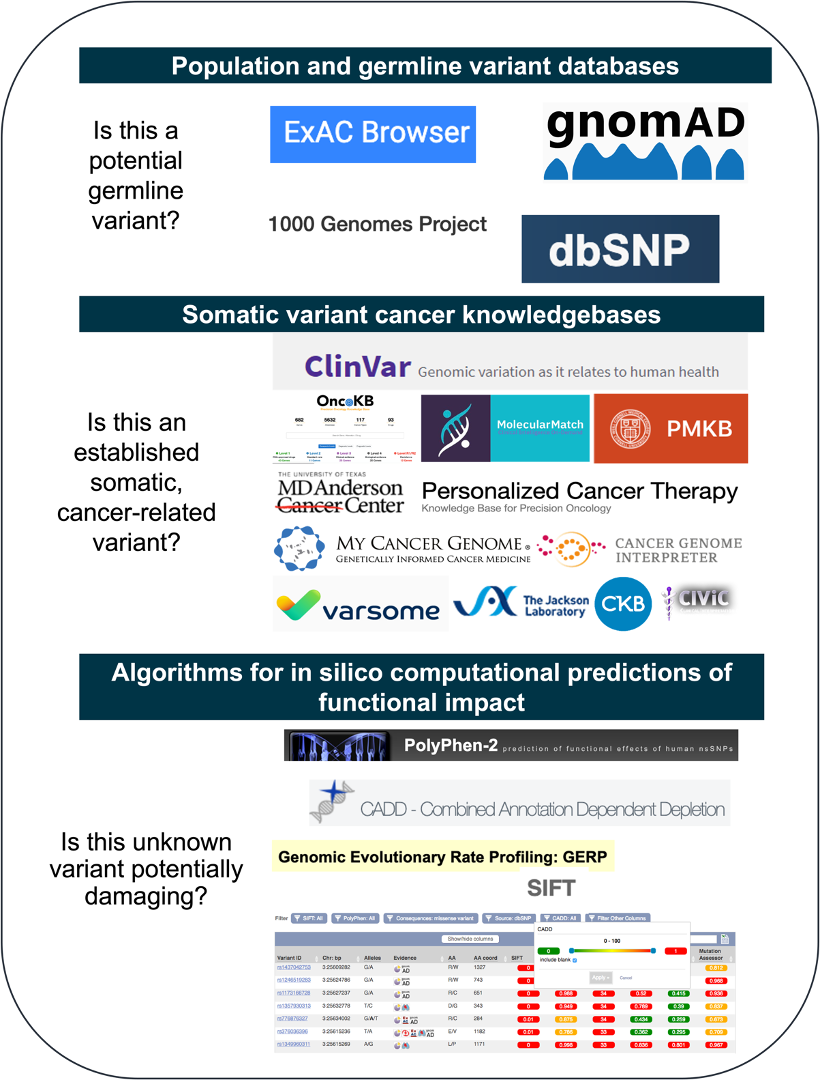
For example, the following questions must be answered when interpreting copy number or mutation data:
Answering these questions requires a careful, comprehensive interpretation process. Molecular geneticists and genome scientists have the experience to mine the various publicly available variant databases to derive conclusions about potential germline variants, pathogenicity, and clinical actionability. Ultimately, the goal is to construct a report for oncologists with only the most clinically relevant patient-specific information displayed clearly and concisely such that a clinician may quickly and accurately differentiate between actionable targets, resistance markers, and alterations with little or no evidence of clinical relevance.
Combining variant annotation and interpretation with treatment decision-making within the context of a specific tumor type is a complex process. In this regard, several commercial providers have begun to launch a variety of clinical decision support tools that convert the molecular information obtained via NGS into a clinical-grade report describing treatment options and clinical trials matched to the patient's genomic profile. In the future, once standardized, such solutions may streamline genomic interpretation workflows and provide labs with the opportunity to scale their services, as they eliminate the manual labor associated with deriving clinical evidence from cancer knowledgebases, drug regulatory agencies, published literature, and clinical trial registries.
Example clinical decision support tools:
Relevant literature evaluating such tertiary NGS analysis tools and oncogenomic reporting:
| Literature |
|---|
| Katsoulakis E, Duffy JE, Hintze B, Spector NL, Kelley MJ. Comparison of Annotation Services for Next-Generation Sequencing in a Large-Scale Precision Oncology Program. JCO Precis Oncol. 2020;4. https://doi.org/10.1200/PO.19.00118. eCollection 2020. |
| Perakis SO, Weber S, Zhou Q, Graf R, Hojas S, Riedl JM, et al. Comparison of three commercial decision support platforms for matching of next-generation sequencing results with therapies in patients with cancer. ESMO Open. 2020;5(5):e000872–2020–000872. |
| Yaung SJ, Krishna S, Xi L, Ju C, Palma JF, Schmid M. Assessment of a Highly Curated Somatic Oncology Database to Aid in the Interpretation of Clinically Important Variants in Next-Generation Sequencing Results. J Mol Diagn. 2020 Nov;22(11):1356-1366. doi: 10.1016/j.jmoldx.2020.08.004. Epub 2020 Sep 19. PMID: 32961319. |
| Boichard A, Richard SB, Kurzrock R. The Crossroads of Precision Medicine and Therapeutic Decision-Making: Use of an Analytical Computational Platform to Predict Response to Cancer Treatments. Cancers (Basel). 2020 Jan 9;12(1):166. doi: 10.3390/cancers12010166. PMID: 31936627; PMCID: PMC7017109. |
| Yaung SJ, Pek A. From Information Overload to Actionable Insights: Digital Solutions for Interpreting Cancer Variants from Genomic Testing. Journal of Molecular Pathology. 2021; 2(4):312-318. https://doi.org/10.3390/jmp2040027 |
| Wagner AH, Walsh B, Mayfield G, Tamborero D, Sonkin D, Krysiak K, et al. A harmonized meta-knowledgebase of clinical interpretations of somatic genomic variants in cancer. Nat Genet. 2020;52(4):448–57. |
| Reisle, C., Williamson, L.M., Pleasance, E. et al. A platform for oncogenomic reporting and interpretation. Nat Commun13, 756 (2022). https://doi.org/10.1038/s41467-022-28348-y |
However, it is impossible to remove the need for expert human insight for the interpretation of molecular profiling results within the context of the individual patient. As the frontline leaders of precision cancer medicine, oncologists must ultimately make the final decision based on all available evidence as to which treatment plan best suits his/her patient. In this regard, the concept of the molecular tumor board (MTB) has demonstrated its importance in standardizing the processing of patient history, clinical data, and NGS profiling results for treatment decision-making. With an MTB infrastructure, clinics can harvest the diverse expertise from oncologists, pathologists, geneticists, variant scientists, bioinformaticians and clinical trial coordinators to ensure a reproducible yet personalized analysis workflow for the patient in question, not to mention devise innovative research approaches.
This was just a taste. Like what you see? Subscribe for deeper dives with programs tailored to the following topics:
- Biology of cell-free DNA (release, clearance, half-life, detection levels across tumor entities, etc.)
- Pre-analytics (choosing a blood collection tube, centrifugation, storage, extraction, quantification)
- Library preparation approaches
- Sequencing approaches
- Intro to bioinformatics basics
- Analysis approaches to detection of: tumor fraction, fragment size, SCNAs, mutations, epigenetic alterations and open chromatin
- Fragmentomics applications
- Interpretation of data from liquid biopsy
- Clinical decision support tools
- Breakdown of more individual case studies
Start now to make liquid biopsy a routine tool in your precision medicine portfolio. Click below to subscribe to our content.
We are a precision oncology community whose goal it is to make state-of-the-art personalized care available to the majority of patients.
It's our aim to educate patients and enable clinicians to implement state-of-the-art precision oncology approaches throughout the entire patient journey, driving the adoption of precision medicine by hospitals, payers and providers.
Strengthen your understanding of the molecular profiling data generated by the latest next-generation sequencing technology that informs your patient care right here at Vessel.